We arrived in Kisumu the day before World Malaria Day. Unfortunately this marked the end of the journey for Isaac, our project medic. Isaac had provided invaluable help with planning the trip, particularly with regards to our health and safety and risk management, and if we all get to the end of the trip without malaria it will be in large part thanks to Isaac’s insistence on performing his own ‘indoor residual spraying’ campaign with a can of Raid at every place we stayed. He was to return to the UK for his sister’s wedding so we had a final meal in Kisumu and Isaac gamely and beautifully, sang karaoke on his last night (Adele’s Someone Like You, if you’re wondering).
On WMD itself, we were fortunate to be invited to the main Kenyan commemoration event which was happening in Siaya, a short drive from Kisumu. Siaya had been chosen as it’s one of the most endemic malarial counties in Kenya, which are all in the region of Western Kenya around Lake Victoria.
In addition to the main event, there was to be an unveiling of the new Kenya Malaria Strategy (KMS) for the next five years. We arrived just in time to hear the Head of the National Malaria Elimination Centre, Dr Waqo Erjesa, present the new plan that has the ambitious goal of reducing malaria incidence and deaths across Kenya to at least 75% of 2016 levels by 2023. The KMS is based on six main objectives, ranging from the continued use of malaria preventative interventions (bednets, spraying, larval source management and preventative treatment in pregnancy) to case management, community engagement, systems development and surveillance.
Two things stood out for me. The first was that, although a succession of partners spoke after Dr Waqo, including representatives from the US government, the UN family, and the local governor, there was only once a mention of needing to think about importation and of cross border collaboration. Although this is certainly on the agenda, it is much further down the priority list than the regional collaboration model of Elimination8 that we had heard so much about in Namibia and Zambia.
The second was the repeated use of the term sustainability. There is clear ambition within Kenya to move to a situation where there is less reliance on donor aid and outside expertise to help with the fight against malaria. This sentiment was echoed in an article by Dr Fredros Okumu, Director of Science at Ifakara Health Institute in Tanzania, which calls for African governments to ‘build resilience’ from within their borders, rather than rely on the short term gains of buying in malaria intervention commodities from outside.
Understanding the importance of malaria vectors
Back in Kisumu, we visited the lab of Dr Eric Ochomo, our main collaborator here and Head of Entomology at the KEMRI Centre for Global Health Research. Eric and his team perform a range of molecular analyses on mosquito populations, from assaying resistance to understanding the species composition of different mosquito populations. They are also busy monitoring mosquito populations during the trials of several new insecticide-based interventions. Despite our focus so far on the parasite, it’s important not to understate the importance of vector control. A large part of the reduction in malaria burden over the first decade and a half of the millennium was down to increased adoption of bednets and more widespread use of indoor residual spraying, both interventions that target the mosquito. If you can stop the vector, you can go a long way to controlling the disease. Indeed, given the importance of vector control and because insecticide resistance has already emerged, the loss of vector control tools is potentially a greater short term threat to malaria control across Africa than antimalarial drug resistance in the parasite.
Much like our time in Zambia, we were going to work with Eric and his team for about 10 days, so we spent a bit of time planning our work. As well as our desire to train the team, we were hopeful that we’d be able to deploy the mobile lab in the field at least once during our stay, but were going to take the lead from Eric as to what the best approach would be. In anticipation of our time at KEMRI, Eric, Isaac and I had submitted and obtained ethical approval to collect mosquitoes for sequencing, so we decided that after an initial trial of the MinION at the lab in KEMRI, we’d visit Teso, a village near to the Uganda border where Eric has an ongoing project.
On Saturday eight of us travelled up to Busia, a team including two ‘mosquito catchers’, Lapan and Mush, three students, Diana, Matthew and Steve, and Eric, Jason and myself. The next day we were up early to drive the 30 minutes from Busia to Teso and once we arrived, having for the first time on the trip momentarily put the car into four wheel drive, we parked up at the edge of the village and went to speak to the elders.
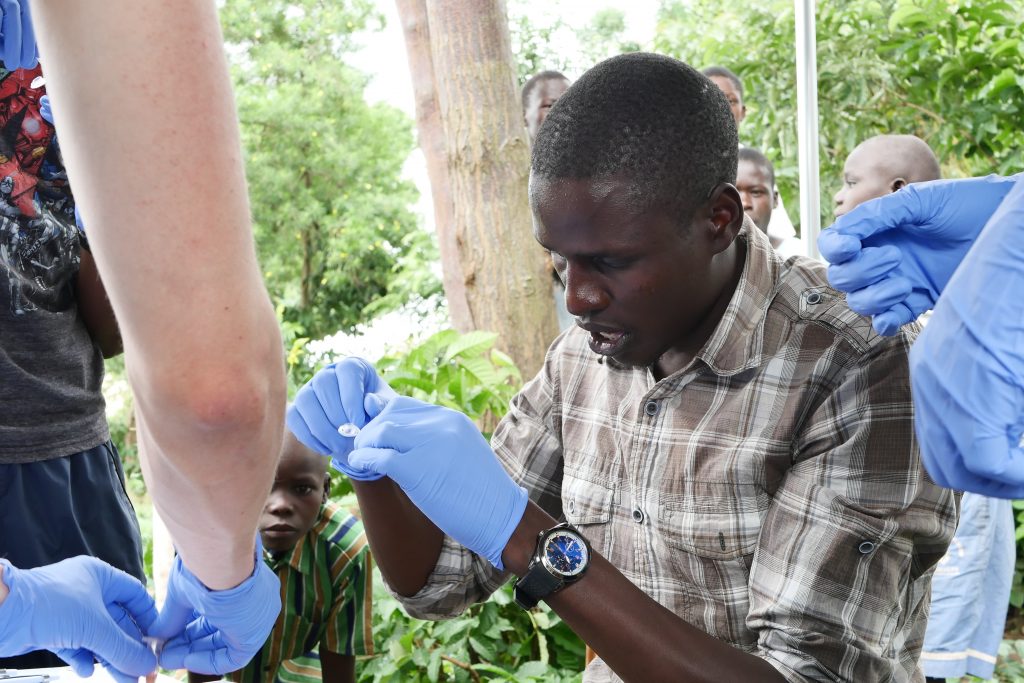
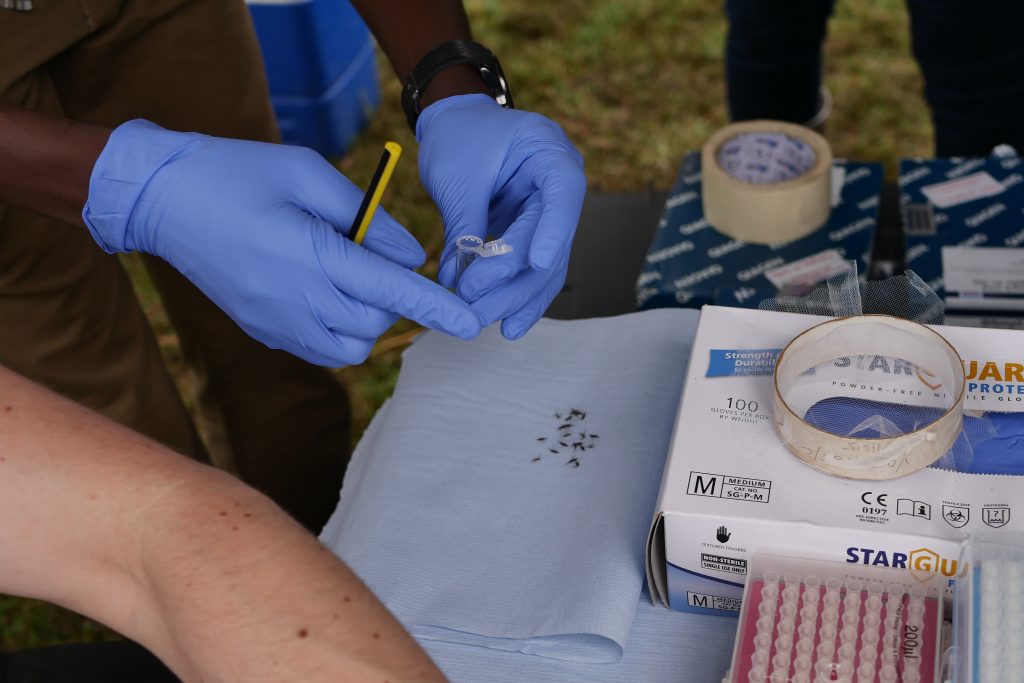
Our introductions having been made, Lapan and Mush enlisted one of the senior men to help catch some mosquitoes. The collection process here involves visiting each of the houses in the village with an aspirator – a sort of hoover with filter that gently sucks insects into a cup – and sucking in any unsuspecting insects residing around the roofs, walls and floors of houses. The mosquito catchers were a slick team and could tell by sight not only which species of mosquito were there (to a first approximation) but also which sex they were (males have fluffy antennae). Only female Anopheles mosquitoes bite, so using a pooter they discard the males and anything that was not Anopheline. It was fascinating to experience mosquito collection and it made me think about my colleagues back in Oxford who work on mosquito genetics and how many of those samples will have originally been collected in this way.
With the mosquitoes, we wanted to test a similar protocol to the one tried on parasite DNA, that is we wanted to amplify specific genes from a few samples and run these on the machine. In the case of parasite DNA, these were known antimalarial resistance genes and of equivalent interest for mosquitoes are insecticide resistance genes. Unfortunately, we couldn’t get our gene-specific assays to work (sometimes labwork can feel more like art than science!) so we instead opted to sequence the full genome of ten samples. This would allows us to trial the feasibility of the lab and would give us plenty of data to work through with the team, but would have less immediate practical utility. To put it another way, we would still be able to sequence genes associated with insecticide resistance, but would also be sequencing all of the rest of the genome as well. We’d have plenty of data to play with, even if most of it has little to do with insecticide resistance. Eric’s team are interested in developing the portable sequencing protocols to test many hundreds of individual samples in a single run, and in this case, looking at a few key parts of the genome rather than the whole thing would be beneficial.
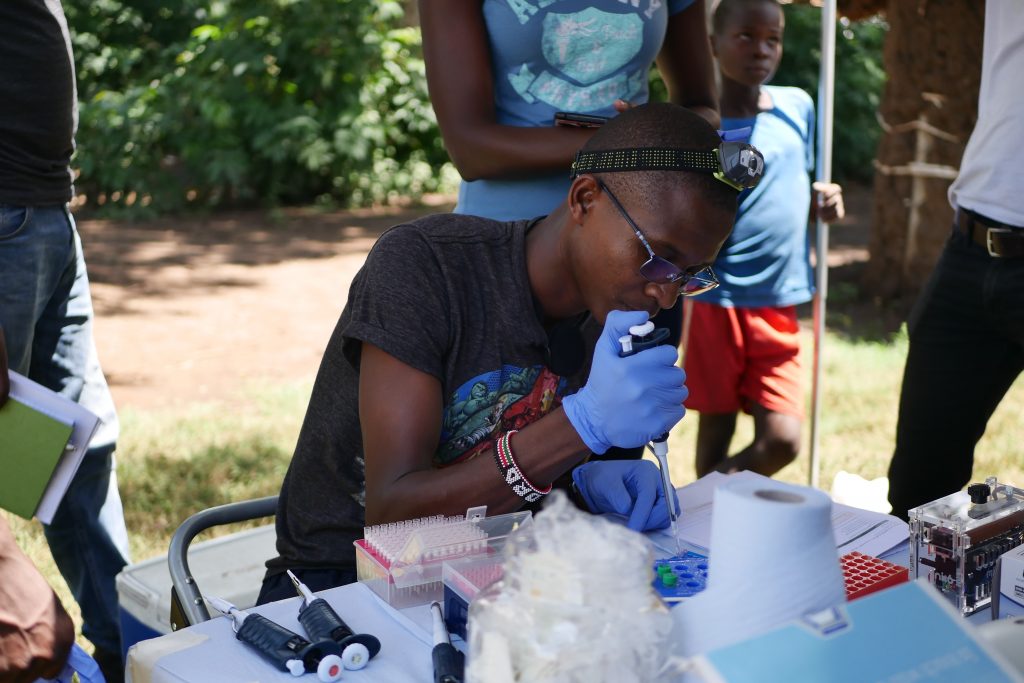
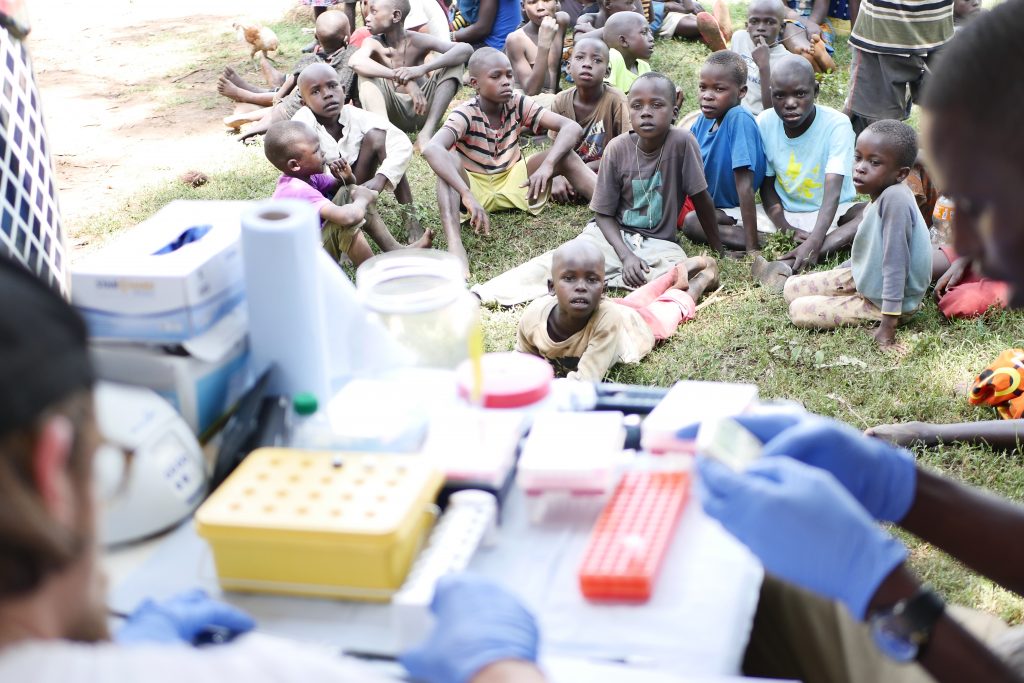
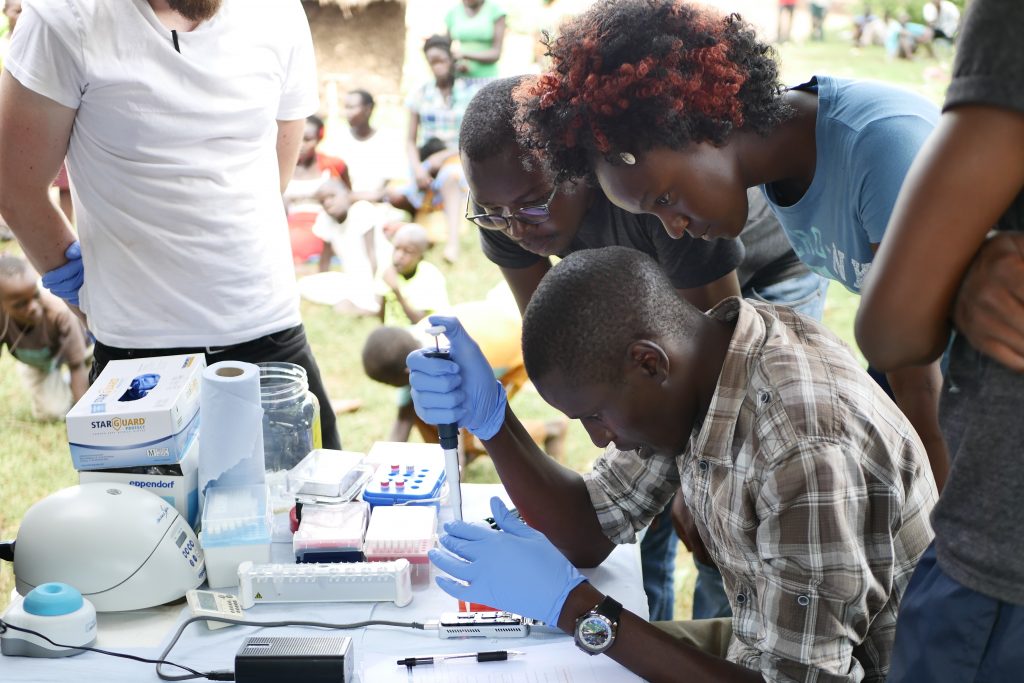
Once the first few Anopheles mosquitoes started to come in we set up the field lab. Jason was going to lead three masters students Diana, Matthew and Steve through the sequencing protocol. The first thing to do was extract DNA from the mosquitoes, so the team got busy crushing up mosquitoes in tubes before adding some reagents and spinning them down. After this were a few preparation steps, each of which takes an hour or so. We had had an audience from the moment we arrived so tried to explain what we were doing and why. Everyone was pretty excited by the car and the lab and were interested to see what we were up to.
As the day wore on the sun came out and it started to get very warm. We’d pulled the awnings of the car out for protection from the fierce equatorial sun. (The day before we had to cross the equator, and for the first time since we’d got to Namibia almost six weeks ago we were back in the northern hemisphere, albeit temporarily.) By just after 2pm, 5 hours after the extractions had started, Matthew was ready to load the MinION. We all looked on as Matthew dropped the DNA that had originated from mosquitoes which had still been living that morning, onto the MInION. They pressed go, the machine started to churn and within a few moments, it was reading the DNA sequence.
What a truly incredible experience to be sequencing DNA in a village, without external electricity, with the vast majority of the work being done by our colleagues from KEMRI, with only a couple of days’ training.
Whilst the use case for sequencing ‘out of the back of a Land Rover’ remains to be made, and I’m the first to admit that it wasn’t necessary in this case to do the sequencing there and then, in my opinion this day demonstrates three incredibly important lessons.
The first is that genetic data generation, with the application of machines like the Oxford Nanopore MinION, can be done anytime anywhere. Historically, DNA has been sequenced in large sequencing facilities, and whilst this doesn’t in any way suggest a need to replace such centres, as we’ll always need to produce high quality, standardised genomic data, this work adds to the growing cannon of examples that demonstrate the potential for anyone with a bit of training to get one of these machines to work.
Secondly, if Matthew, Diana, and Steve can sequence DNA in these extreme conditions, then people can have the confidence to sequence DNA in any of the hundreds of small research labs spread across Africa, including those back at KEMRI were we have been working this last week. We came with a few potential use cases for portable genetic sequencing, but it’s clear to me that the people we’ve met in Namibia, Zambia and Kenya have their own questions to answer and their own need for genetic sequencing data. If MinIONs can be used to get people into genetic sequencing, from the ground up, and researchers can gain the confidence to use genetics to answer their own questions then genomic science in Africa will be in rude health in years to come. It will grow with scientists who have been able to take ownership of the full process of scientific enquiry, from observation and hypothesis generation to data analysis and conclusion.
Finally, I think and hope that our demonstration will inspire. The team at KEMRI were buzzing about the potential of the technology. No one exemplifies this more than Diana. When we arrived at the KEMRI labs the day after our travails in Teso, she had already run a PCR of some samples and was keen to run them on the machine. These were not mosquito or parasite DNA, but samples from the gut microbiome of resistant and non-resistant mosquitoes. We ran these on the machine and are already able to help her with the preliminary analyses of these data. Now there’s a long way to go to ensure that the technology can generate good quality data, so we should temper all this excitement with a cool splash of realism for the work needed to truly leverage it in the future, but these seeds of optimism must surely grow if we can sustain this work into the future.
And I hope it’s inspiring to others as well. We started the Mobile Malaria Project with the idea of travelling to Africa to work with malaria geneticists, to learn about their work and to trial cheap, portable genetic sequencing. Throughout our journey, we’ve been met with nothing but positivity, intrigue and excitement. Taking the time out of our lives in Oxford has given us the opportunity to experience the research world of scientists on the frontline of malaria research and I think people have appreciated our attempts to show them some new technology that is relevant to their work. So I hope that anyone who has an idea to do something a bit different and to get out there and see the world will see that it is an eye opening, refreshing and ultimately humbling learning experience.